|

by F.H.C. Ckrick
Medical Research Council
Laboratory of Molecular Biology
Hills Road, Cambridge, England
(Received 21 August 1968)
from
NationalInstituteForMedicalResearch
Website
|
The general features of
the genetic code are described.
It is considered that
originally only a few amino acids were coded, but that
most of the possible codons
were fairly soon brought
into use.
In subsequent steps additional amino acids
were substituted when they were able to confer a
selective advantage, until eventually the code became
frozen in its present form. |
Introduction
The substance of this paper was originally presented at a meeting of
the British Biophysical Society in London on 20 December
1966.
A very brief account appeared shortly after in a letter to Nature
(Crick, 1967ª). When this manuscript was in its first draft, Dr
Leslie Orgel told me that he had already prepared a draft of a
paper on a related theme.
We therefore decided to publish our two
papers together and have collated them to some extent to avoid
overlap. We have not done this for all passages in the two papers
which touch on the same topic, preferring on occasions to let our
slightly different points of view be expressed as differences in
treatment and emphasis. However, broadly speaking, each of us agrees
with the opinion expressed by the other.
Since this paper was originally drafted a very full discussion has
appeared in Carl Woese’s book
The Genetic Code, which should be
consulted for a fuller discussion of many of the points touched on
here.
The Structure
of the Present Genetic Code
The structure of the genetic code is now fairly well known.
The code
is a non-overlapping triplet code. Most, but not all, of the 64
triplets stand for one or another of the 20 amino acids and, in most
cases, each amino acid is represented by more than one codon. The
best present version of the code is shown in Table 1. This is taken
from the 1966 Cold Spring Harbor Symposium on The Genetic Code,
to which the reader is referred as a source of references for many
of the topics discussed here.
Before starting on a detailed examination of this Table a few words
of caution are necessary Although the code shown there has been
mainly derived from studies on Escherichia coli, it must be very
similar in such widely different organisms as tobacco plants and
man. In what follows I shall assume, for convenience of exposition,
that it is identical in all organisms, which is very far from being
proved.
In fact, it is probably untrue for the starting codons.
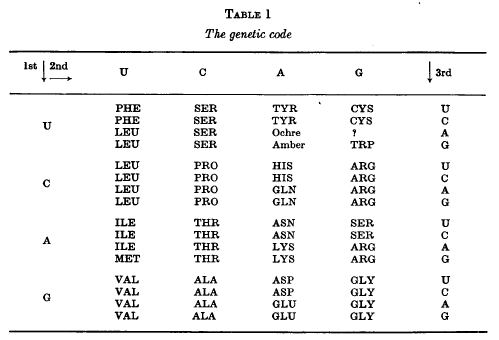
This Table shows the “best
allocations” of the 64 codons at the time of the Symposium. Some
of these allocations are less certain than others. The two
codons marked ochre and amber are believed to signal the
termination of the polypeptide chain. The codons suspected of
being concerned with chain initiation are not indicated here
Again the function of the three presumed
“nonsense” triplets is not known for certain. It is presumed that
UAA (ochre) and UAG (amber) are signals for chain termination and
probably UGA as well, at least in bacteria.
In E. coli there appears to be a special mechanism for initiating
the polypeptide chain, involving formylmethionine and the codons AUG
and GUG. The mechanism in higher organisms (if indeed a special one
exists) is unknown.
Finally, it is uncertain whether there are ambiguous codons; that
is, codons which represent more than one amino acid. Of course, it
is known that mutations can produce errors in the translation
mechanism and so make certain codons ambiguous, but it is not known
whether ambiguity occurs “normally”. Again in what follows I shall
assume that this is not usually the case for present-day organisms.
The basic reason why one can ignore these complications and
uncertainties for the moment is that the broad features of the
genetic code are not likely to be greatly affected by them. What,
then, are the properties of the code which require explanation?
There are some features which are of such a general type that they
do not depend at all upon the details of the code. They are:
(1) there are 4 distinct bases in
the mRNA
(2) each codon is a triplet of bases
(3) only 20 of the numerous possible amino acids are used. In
examining Table 1, however, one is apt to take all these
characteristics for granted. What, then, is special about the
actual details of the genetic code ?
(4) The 20 amino acids are not
distributed at random among the 64 triplets
In fact, several rules can easily be deduced from the Table. For
example,
(a) XYU and XYC always code the
same amino acid.
(b) XYA and XYG often code the same amino acid. The rare
amino acids, methionine and tryptophan, which have only one
codon each, appear to be exceptions to this rule.
(c) In half the cases (8 out of 16) XY represents a single
amino acid, where the XY implies that all four bases are
possible.
(d) In most cases the codons representing a single amino
acid start with the same pair of bases. Thus the two codons
for histidine both start with CA. There are three exceptions
to this:
(e) If the first two bases
consist only of G’s and C’s, then the four codons sharing
the same initial doublet all code the same amino acid. That
is, the meaning of these codons is independent of the third
base. This is in fact true for all codons having C in the
second position. More complicated rules along these lines
can be produced for the remaining codons but they seem to me
to be rather forced.
(5) Even allowing for the grouping
of codons into sets, the amino acids do not seem to be allocated
in a totally random way. For example, all codons with U in the
second place code for hydrophobic amino acids. The basic and
acidic amino acids are all grouped near together towards the
bottom right-hand side of Table 1.
Phenylalanine, tyrosine and
tryptophan all have codons starting with U, and so on. It is
very difficult not to imagine regularities in even a random
grouping but nevertheless the general impression is that
“related” amino acids have to some extent related codons
(Epstein, 1966).
(6) The code is universal (the same in all organisms) or nearly
so.
Why is the Code
Universal?
Two extreme theories may be described to
account for this, though, as we shall see, many intermediate
theories are also possible.
The Stereochemical Theory
This theory states that the code is
universal because it is necessarily the way it is for
stereochemical reasons. Woese has been the main proponent
of this point of view (see Woese, 1967).
That is, it states that
phenylalanine has to be represented by UUJÍ, and by no other
triplets, because in some way phenylalanine is stereochemically
“related” to these two codons.
There are several versions of
this theory. We shall examine these shortly when we come to
consider the experimental evidence for them.
The Frozen Accident Theory
This theory states that the code is
universal because at the present time any change wovld be leihal,
or at least very strongly selected against.
This is because in
all organisms (with the possible exception of certain virases)
the code determines (by reading the mRNA) the amino acid
sequences of so many highly evolved protein molecules that any
change to these would be highly disadvantageous unless
accompanied by many simultaneous mutations to correct the
“mistakes” produced by altering the code.
This accounts for the fact that the code does not change.
To
account for it being the same in all organisms one must assume
that all life evolved from a single organism (more strictly,
from a single closely interbreeding population).
In its extreme
form, the theory implies that the allocation of codons to amino
acids at this point was entirely a matter of “chance”.
The Stereochemical
Theory - Experimental Evidence
In its extreme form, the stereochemical theory states that the
postulated stereochemical interactions are still taking place today.
It should therefore be a simple matter to prove or disprove such
theories.
Pele and Welton (Pele & Welton, 1966; Welton & Pele,
1966) nave suggested from a study of models that there is in many
cases a specific stereochemical fit between the amino acid and the
base sequence of its codon on the appropriate tRNA.
Unfortunately,
their models were all built backwards (Crick, 19676) so their claims
are without support. Such a theory implies that the expected codon
sequence occurs somewhere on each tRNA. For example, no such
sequence occurs in the tRNA for tyrosine either from yeast (Madison,
Everett & King, 1966) or from E. coli (Goodman, Abelson, Landy,
Brenner & Smith, 1968). In our opinion this idea has little chance
of being correct.
A more reasonable idea is that the amino acid fits the anticodon on
the tRNA. At least this has the advantage that it is always present.
A model along these lines for proline has been briefly described by
Dunnill (1966), but so far no detailed description has been
published, nor has he extended his model-building to other amino
acids.
The experimental evidence has already established that when the
activating enzyme transfers the amino acid to the tRNA, the
interaction is not solely with the anticodon and the common... CCA
terminal sequence.
This is shown by the fact that an activating
enzyme from one species will not always recognize the appropriate tRNA from a different species although the anticodons must be very
similar if not identical in different species (for a summary of the
data, see Woese, 1967, p. 125). However, this does not preclude the
idea that the interaction is partly with the anticodon and partly
with some other part of the tRNA.
The best way to disprove the theory (if indeed it is false) would be
to change the anticodon of some tRNA molecule and show that
nevertheless it accepted the same amino acid from the activating
enzyme.
This has already been done for the minor tyrosine tRNA of E.
coli whose anticodon has been changed (in an Su + strain) from GUA
to CUA (Goodman et al., 1968) although the experiments need to be
done quantitatively. Further examples of such changes are likely to
be reported in the near future.
Until this is done we must reserve
final judgment on the amino acid-anticodon interaction theory; but
we consider it unlikely to be correct, except per-haps in a few
special cases.
Even if it were established that the activating enzyme recognizes
the anticodon, this would not by itself prove that the recognition
is done by inserting the amino acid in a cage formed by the
anticodon. Notice that the activating enzyme would have to release
amino acid from its own recognition cavity and then insert it into
the recognition site on the tRNA.
Moreover, when the amino acid has
been transferred to the tRNA and the activating enzyme has diffused
elsewhere, the amino acid could not stay in the anticodon cage
without blocking the interaction with the codon on the mRNA. None of
this is impossible but it is certainly elaborate.
It is not easy to see at this stage what evidence would be needed to
prove that the anticodon does indeed form a cage for the amino acid,
though if the tRNA (or perhaps a fragment of it) could be
crystallized it might be possible to see the amino acid sitting in
such a position.
The present experimental evidence, then, makes it unlikely that
every amino acid interacts stereochemically with either its codon or
its anticodon. It by no means precludes the possibility that some
amino acids interact in either of these ways, or that such
interactions, even though now not used, may have been important in
the past, at least for a few amino acids.
We must now leave the
system as it is today and turn to the examination of primitive
systems.
The Primitive
System
It is almost impossible to discuss the origin of the code without
discussing the origin of the actual biochemical mechanisms of
protein synthesis.
This is very difficult to do, for two reasons: it
is complex and many of its details are not yet understood.
Nevertheless, we shall have to present a tentative scheme, otherwise
no discussion is possible.
In looking at the present-day components of the mechanism of protein
synthesis, one is struck by the considerable involvement of
non-informational nucleic acid. The ribosomes are mainly made from
RNA and the adaptor molecules (tRNA) are exclusively RNA, although
modified to contain many unusual bases. Why is this?
One plausible explanation, especially
for rRNA, is that RNA is “cheaper” to make than protein. If a
ribosome were made exclusively of protein the cell would need more
ribosomes (to make the extra proteins, which would not be a
negligible fraction of all the proteins in the cell) and thus could
only replicate more slowly.
Even though this may be true, we cannot
help feeling that the more significant reason for rRNA and tRNA is
that they were part of the primitive machinery for protein
synthesis.
Granted this, one could explain why
their job was not taken over by protein, since
(i) for rRNA, it would be too
expensive
(ii) for tRNA, protein may not be able to do such a neat job
in such a small space
In fact, as has been remarked elsewhere,
tRNA looks like Nature’s attempt to make RNA do the job of a protein
(Crick, 1966).
If indeed rRNA and tRNA were essential parts of the primitive
machinery, one naturally asks how much protein, if any, was then
needed. It is tempting to wonder if the primitive ribosome could
have been made entirely of RNA.
Some parts of the structure, for
example the presumed polymerase, may now be protein, having been
replaced because a protein could do the job with greater precision.
Other parts may not have been necessary then, since primitive
protein synthesis may have been rather inefficient and inaccurate.
Without a more detailed knowledge of the structure of present-day ribosomes it is difficult to make an informed guess.
It is not too difficult to imagine that the early tRNA molecules had
no modified bases (so that no modifying enzymes were needed), but it
is much more difficult to decide whether activating enzymes were
then essential.
An attractive idea (suggested to us by Dr Oliver
Smithies) is that the primitive tRNA was its own activating
enzyme. That is, that its structure had a cavity in it which
specifically held the side-chain of the appropriate amino acid in
such a position that the carboxyl group could be easily joined on to
the terminal ribose of the tRNA.
It is thus not impossible to imagine that the primitive machinery
had no protein at all and consisted entirely of RNA. This is
discussed at much greater length in the companion paper by Dr L.
E. Orgel, where the importance of the ease of replication of
nucleic acid is emphasized.
We are faced with the question of the
origin of all this RNA. Could the appropriate sequences have arisen
by chance?
We do not feel this is totally
impossible, for three reasons.
(a) Some natural catalyst (such as a
mineral) for random nucleotide polymerization may exist. If this
were so, RNA may have been made at very many places on the
earth’s surface over a very considerable period of time, so that
altogether an enormous number of different sequences may have
been synthesized. It is difficult to assess the value of this
idea, since such a natural catalyst has not yet been discovered.
Another possibility is that a crude template mechanism developed
at an early stage. This is fully discussed in the companion
paper.
(b) The mechanism of “random” synthesis may preferentially
produce structures with multiple loops (this is also discussed
in the companion paper) so that sequences of this sort (which
are indeed found in tRNA and rRNA) may have been synthesized
preferentially. Moreover, the actual base-pairs used in the
base-paired regions may not be critical for their structures. In
short, the synthesis of an acceptable rRNA and tRNA may not have
been so unlikely as it seems at first sight.
(c) The base-sequences needed may have been repetitive. For
example, the early tRNA molecules may have been very alike, only
differing in the anticodon and in the region of the presumed
cavity. For all we know, the structure of the large rRNA
molecules may have been partly repetitive. These repetitions
might have been produced rather easily if there were an RNA
replicase available. Possibly the first “enzyme” was an RNA
molecule with replicase properties. Thus a system based mainly
on RNA is not impossible. Such a system could then start to
synthesize protein and thus could evolve very rapidly by natural
selection. We shall not discuss here the difficult problem of
how the various components were kept together, that is, the
origin of a cell.
The point of this sketch is to impress
the reader with the great difficulty of the problem.
It would
certainly be easier if specific stereo chemical interactions could
occur between amino acids and triplets of bases, but even if these
are possible the origin of the present ribosomal translation
mechanism presents grave difficulties.
The Primitive
Code
We must now tackle the nature of the primitive code and the manner
in which it evolved into the present code.
It might be argued that the primitive code was not a triplet code
but that originally the bases were read one at a time (giving 4
codons), then two at a time (giving 16 codons) and only later
evolved to the present triplet code. This seems highly unlikely,
since it violates the Principle of Continuity.
A change in codon size necessarily makes
nonsense of all previous messages and would almost certainly be
lethal. This is quite different from the idea that the primitive
code was a triplet code (in the sense that the reading mechanism
moved along three bases at each step) but that only, say, the first
two bases were read. This is not at all implausible.
The next general point about the primitive code is that it seems
likely that only a few amino acids were involved. There are several
reasons for this. It certainly seems unlikely that all the present
amino acids were easily available at the time the code started.
Certainly tryptophan and methionine look like later additions.
Exactly which amino acids were then
common is not yet clear, though most lists would include glycine,
alanine, serine and aspartic acid.
However, if sterepchemical
interaction played a part in the primitive code, this might select
amino acids which were available but not particularly common. Again,
it seems unlikely that the primitive code could code specifically
for more than a few amino acids, since this would make the origin of
the system terribly complicated.
However, as Woese (1965) has
pointed out, the primitive system might have used classes of amino
acids. For example, only the middle base of the triplet may have
been recognized, a U in that position standing for any of a number
of hydrophobic amino acids, an A for an acidic one, etc.
Even though few amino acids (or groups of amino acids) were
recognized, it seems likely that not too many nonsense codons
existed, otherwise any message would have had too many gaps. There
are various ways out of this dilemma. For example, as mentioned
above, only one base of the triplet might have been recognized.
Another possibility, however, is that the early message consisted
not of the present four bases, but perhaps only two of them.
The Number of
Bases in the Primitive Nucleic Acid
The only strong requirements for the primitive nucleic acid is that
it should have been easy to replicate, and that it should have
consisted of more than one base, otherwise it could not carry any
information in its base sequence.
One cannot even rule out the
possibility that the base sequence of the two chains was
complementary (as in the present DNA). Perhaps a structure is
possible with only two bases in which the two chains run parallel
(rather than anti-parallel) and pairing is like-with-like. It would
certainly be of great interest if such a structure could be
demonstrated experimentally.
Leaving this possibility on one side and restricting ourselves to
complementary structures, we see that the number of bases must be
even. If there were only two in the primitive DNA, the question
arises as to which two. The obvious choices are either A with U (or
T) or G with C. A less obvious possibility (suggested some time ago
by Dr Leslie Orgel, personal communication) is A with I
(where I stands for inosine, having the base hypoxanthine).
It is not certain that a double helix
can be formed having a random sequence of A’s and I’s on one chain
and the complementary sequence (dictated by A-I or I-A pairs) on the
other chain, but it is not improbable, especially as the RNA
polymers poly A and poly I can form a double helix.
Several advantages could be claimed for this scheme. Adenine is
likely to be the commonest base available in the primitive soup, and
inosine could arise from it by deamination.
Thus the supply of
precursors might be easier than in the case of the other two
alternatives, though how true this is remains to be established.
Then again in a random (A, I) sequence I would presumably code in
the same way as G does now, at any rate for the first two positions
of the triplet.
If we can use the present code as a
guide (though we shall argue later that this may be misleading), it
is noticeable that the triplets containing only A’s or G’s in their
first two bases (the bottom right-hand corner of the Table) do
indeed code for some of the more obviously primitive amino acids.
It is important to notice that a scheme of this sort (or even one
with like-with-like pairing) does not violate the principle of
continuity. To change over from an (A, I) double helix to one like
the present one but having A, I, U and C, the only steps required
are a change in the replicase to select smaller base-pairs, and a
supply of the two new precursors. The message carried (by the “oíd”
chain) is unaltered by this step.
Gradually mutations would produce U’s
and C’s on this chain and the new codons thus produced could be
brought into use as the mechanism for protein synthesis evolved.
Eventually G would be substituted for I. At no stage would the
message become complete nonsense. The idea that the initial nucleic
acid contained only two bases is thus a very plausible one.
It
remains to be seen whether primitive ribosomal RNA and primitive tRNA could be constructed using only two bases.
The
Stereochemical Alternative
As stated earlier, it seems very unlikely that there is any
stereochemical relationship between all the present amino acids and
specific triplets of bases; but it is by no means ruled out that a
few amino acids can interact in this way.
If this were possible, it
would certainly help in the initial stages of the evolution of the
code.
However, sooner or later a transition
would have had to be made to the present type of system, involving
tRNA’s, ribosomes, etc. It seems to us that this could only happen
easily if the code at that stage was fairly simple and only coded a
rather small number of amino acids.
The Evolution of the
Primitive Code
Whatever the early steps in the evolution of the code, it seems
highly likely that it went through a stage when only a few amino
acids were coded.
At this stage either the mechanism was rather
imprecise and thus could recognize most of the triplets, or only a
few triplets were used, perhaps because the message contained only
two types of base. We must now consider what would happen next.
A complication should be introduced into this simple picture. It
could well be that at this stage the recognition mechanisms were not
very precise and that any given codon corresponded to a group of
amino acids (see Woese, 1965, who has stressed this point).
Thus codons for alanine might also
incorporate glycine, those for threonine might also code serine,
etc. However, it is by no means certain that this happened. It seems
highly likely that a “cavity” to accept threonine would also accept
serine to some extent, but the converse mistake is less likely and
could depend on the exact nature of the structure involved. Thus,
though the early coding machinery probably produced errors, we can
only guess at their extent.
We shall argue that by far the most likely step was that these
primitive amino acids spread all over the code until almost all the
triplets represented one or other of them.
Our reasons for believing
this are that too many nonsense triplets would certainly be selected
against, so that most codons would quickly be brought into use (Sonneborn,
1965). In addition, it would be easier to produce a new tRNA,
altered only in its anticodon, while still recognizing the amino
acid, than to produce both a new anticodon and a new recognition
system for attaching a new amino acid.
Thus, we can reasonably expect that the
intermediate code had two properties:
(i) few amino acids were
coded, and
(ii) almost all the triplets
could be read.
Moreover, because of the way this
primitive code originated, the triplets standing for any one amino
acid are likely to be related.
At this stage the organism could only
produce rather crudely made protein, since the number of amino acids
it could use was small and the proteins had probably not evolved
very extensively.
The final steps in the evolution of the code would involve an
increase in the precision of recognition and the introduction of new
amino acids. The cell would have to produce a new tRNA and a new
activating enzyme to handle any new amino acid, or any minor amino
acid already incorporated because of errors of recognition.
This new tRNA would recognize certain triplets which were probably already
being used for an existing amino acid. If so, these triplets would
be ambiguous.
To succeed, two conditions would have to
be fulfilled.
(1) The new amino acid should not
upset too much the proteins into which it was incorporated. This
upset is least likely to happen if the old and the new amino
acids are related.
(2) The new amino acid should be a positive advantage to the
cell in at least one protein. This advantage should be greater
than the disadvantages of introducing it elsewhere.
In short, the introduction of the new
amino acid should, on balance, give the cell a reproductive
advantage.
For the change to be Consolidated we would expect many further
mutations, replacing the ambiguous codons by other codons for the
earlier amino acid when this was somewhat better for a protein than
the later one. Thus, eventually the codons involved would cease to
be ambiguous and would code only for the new amino acid.
There are several reasons why one might expect such a substitution
of one amino acid for another to take place between structurally
similar amino acids.
-
First, as mentioned above, such a
resemblance would diminish the bad effects of the initial
substitution.
-
Second, the new tRNA would probably
start as a gene duplication of the existing tRNA for those
codons.
Moreover, the new activating enzyme
might well be a modification of the existing activating enzyme.
This
again might be easier if the amino acids were related. Thus, the net
effect of a whole series of such changes would be that similar amino
acids would tend to have similar codons, which is just what we
observe in the present code.
It is clear that such a mechanism for the introduction of new amino
acids could only succeed if the genetic message of the cell coded
for only a small number of proteins and especially proteins which
were somewhat crudely constructed. As the process proceeded and the
organism developed, more and more proteins would be coded and their
design would become more sophisticated until eventually one would
reach a point where no new amino acid could be introduced without
disrupting too many proteins.
At this stage the code would be frozen.
Notice that it does not necessarily follow that the original codons,
of the original primitive code (as opposed to the intermediate code)
will necessarily keep their assignments to the primitive amino
acids. In other words, the evolution of the code may well have wiped
out all trace of the primitive code.
For this reason arguments about
which base-pair carne into use first on the nucleic acid should not
depend too heavily on the assignments of the present code.
The idea described above is crucial to
the evolution of the code. It seems to me not to be the same as the
idea, suggested by several authors (Sonneborn, 1965; Goldberg
& Wittes, 1966), that the code is designed to minimize the
effects of mutations. The implication is that the mutations are
those occurring in the many proteins of the organism, and in fact
are still occurring today.
This is not quite the same as the idea
that it is the situation produced by the introduction of a new amino
acid to the developing code that we have to consider. Moreover, the
disturbances had to be minimized not to the present day proteins but
to the small number of more primitive proteins then existing.
The
minimizing of the effects of mutations is in any case likely to have
only a small selective advantage even at the present time, and I
think it unlikely that it could have had any appreciable effect in
molding the genetic code. Woese (1967) has made the same
point.
An idea rather close to the one presented above has been developed
by Woese (1965). He emphasizes in his discussion the fact
that the early translation mechanism would probably be prone to
errors. This is indeed an important idea and may well be what
actually occurred but it is not identical to the idea suggested
above, as can be easily seen by making the rather unlikely
assumption that the early mechanism was rather accurate.
In this case Woese’s ideas are
irrelevant and one is driven to the scheme outlined above.
Nevertheless, Woese’s discussion (Woese, 1967) follows much
the same line as that presented here. However, he argues that by
this mechanism it is unlikely that the code could reach the truly
optimum code.
There is no reason to believe, however,
that the present code is the best possible, and it could have easily
reached its present form by a sequence of happy accidents. In other
words, it may not be the result of trying all possible codes and
selecting the best. Instead, it may be frozen at a local minimum
which it has reached by a rather random path.
On the other hand, the basic idea has been very clearly stated by
Thomas H. Jukes (1966) in his book
Molecules and Evolution (p. 70)
though he does not give it any particular emphasis.
There is one feature of the process by which new amino acids were
added to a primitive code which is far from clear. This is why
several versions of the genetic code did not emerge. It is, of
course, easy to say that in fact several did emerge and only the
best one survived, but the argument is rather glib. A detailed
discussion of what was likely to have happened at this period would
involve the consideration of genetic recombination.
Did it occur at a very early stage,
perhaps even before the evolution of the cell, and, if so, what form
did it take? Surprisingly enough, no writer on the evolution of the
code seems to have raised this point. Naturally only rather simple
processes would be expected, but the selective advantages of such a
process would be very great. Perhaps a simple fusion process would
suffice for the origin of the code (a suggestion made by Dr
Sydney Brenner, personal communication).
This would provide spare genes for
further evolution and in as far as the code for the fusing organisms
differed it would produce fruitful ambiguities.
One might even argue
that the population which defeated all its rivals and survived was
the one which first evolved sex, a curious twist to the myth of
the Garden of Eden.
General
Features of the Code
We must now go back and ask whether we can explain the general
features of the code in terms of the ideas sketched above.
The Four Distinct Bases
We have argued that
originally there may have been only two bases in the nucleic
acid. Why should there be four today ? The likely answer seems
to be that four were stereochemically possible (i.e. could fit
into a double-helical structure) and that two was too
restrictive a number.
If only the first two bases of the
triplet were originally distinguished, the mechanism could only
code for four things (three amino acids and a space?), and even
if the present “wobble” mechanism applied only a maximum of
eight things could be coded. This could well be too few to
construct really efficient proteins.
Whether six distinct base-pairs are stereochemically possible
has been discussed elsewhere (Rich, 1962; Crick, 1964). It
should be possible to settle this point experimentally.
Why a Triplet?
We have argued that the
code must have been basically a triplet code from a very early
stage, so that one is not entitled to use sophisticated
arguments which would apply only to a later stage, although one
could argue that early organisms with doublet or quadruplet
codes actually existed but became extinct, only the triplet code
surviving.
However, we are inclined to suspect that the reason in this case
may be a structural ‘ one. If indeed there is no direct
stereochemical relationship between an amino acid and a triplet,
the problem of constructing an adaptor to recognize the codon
may be a difficult one to solve.
In effect, one wants to perform a
rather complicated act of recognition within a rather limited
space, since two adaptors need to lie side by side, and attached
to adjacent codons on the mRNA, during the act of synthesis.
This is probably very difficult to perform if protein is used
for the adaptor. On the other hand, nucleic acid, by employing
the base-pairing mechanism, can do a very neat job in a small
space.
For various reasons the adaptor cannot be too simple a molecule.
For example, the amino acids on adjacent adaptors need to be
brought together - this is probably done at the present using the
flexible ... CCA tail. It must have, to some extent, a definite
structure and this is likely to be based on stretches of
double-helix. Thus .the diameter of a double-helix (since two
may have to lie side by side) may have dictated the size of the
codon, in that a doublet-code (moving along two bases at a time)
would present an impossible recognition problem.
The 20 Amino Acids
According to the theory sketched above, both the number 20 and
the actual amino acids in the code are at least in part due to
historical accident.
First note that if the wobble theory of the interaction between
codon and anticodon is correct, then the maximum number of
things which can be coded in a positive way is 32 (say 31 amino
acids and a chain terminator) not 64. Thus, the multiple
representation of eight of the amino acids is not excessive.
On this view, only eight of the 21
things coded appear more than once. If the code evolved as I
have suggested, it would in fact be surprising if each amino
acid did occur only once. However, the theory of wobble must not
be trusted too far, if only because it does not easily explain
the fact that UGA codes differently from both UGUC
and UGG.
Discussion of the actual amino acids used in the code may not be
very profitable. Some less common amino acids, such as cysteine
and histidine, would clearly seem to have an advantage because
of their chemical reactivity; but whether, say, methionine could
be justified in this way seems less obvious. It might be more
useful to consider which amino acids are not used in the code.
However, the answer, if this general
scheme is correct, really depends upon very complicated
considerations, partly accidental, during the early evolution of
the code. In particular, it would depend on the exact nature of
the primitive proteins. It seems unlikely that one could come to
any firm conclusions by following this line of argument.
As already mentioned, the theory does explain in a general way
why similar amino acids often use similar codons. This does not
answer the question whether the allocation of particular amino
acids is entirely due to chance.
However, if it is assumed that the
primitive code used tRNA molecules and that the recognition site
for the amino acid was distinct from the anticodon, then even if
activating enzyme did not exist at this stage and instead the
amino acid fitted into a specific cage in the tRNA, the
association between amino acid and anticodon could be due to
pure chance.
Thus, a code with this property is
not outrageous. Always remember that the present tRNA molecules
must necessarily have evolved at some time or another.
The Two Theories
Contrasted
The evolution of the code sketched here has the property that it
could produce a code in which the actual allocation of amino acid to
codons is mainly accidental and yet related amino acids would be
expected to have related codons.
The theory seems plausible but as a
theory it suffers from a major defect: it is too accommodating.
In a loose sort of way it can explain
anything. A second disadvantage is that the early steps needed to
get the system going seem to require rather a lot of chance effect.
A theory of this sort is not necessarily useless if one can get at
the facts experimentally. Unfortunately, in this problem this is
just what is so difficult to do.
A theory involving stereochemical
relationships between amino acids and triplets, on the other hand,
not only makes it easier to see how the system could start but there
is at least a reasonable chance that well-designed experiments could
prove that such specific interactions are possible. It is therefore
essential to pursue the stereochemical theory.
However, vague models of such
interactions are of little use. What is wanted is direct
experimental proof that these interactions take place (expressed as
binding constants) and some idea of their specificity.
REFERENCES
-
Crick, F. H. C. (1964). In Proc.
Plenary Sessions 6th Int. Gong. Biochem. p. 109. Int. Union
BiocKem. vol. 33. Federation of American Societies for
Experimental Biology.
-
Crick, F. H. C. (1966). Gold Spr.
Harb. Symp. Quant. Biol. 31, 3.
-
Crick, F. H. C. (1967º). Nature,
213, 119.
-
Crick, F. H. C. (19676). Nature,
213, 798.
-
Dunnill, P. (1966). Nature, 210,
1267.
-
Epstein, C. J. (1966). Nature,
210, 25.
-
Goldberg, A. L. & Wittes, R. E.
(1966). Science, 153, 420.
-
Goodman, H. M., Abelson, J.,
Landy, A., Brenner, S. & Smith, J. D. (1968). Nature, 217,
1019.
-
Jukes, T. H. (1966). Molecules
and Evolution. New York: Columbia University Press.
-
Madison, J. T., Everett, G. A. &
King, H. (1966). Science, 153, 531.
-
Pele, S. R. & Welton, M. G. E.
(1966). Nature, 209, 868.
-
Rich, A. (1962). In Horizons in
Biochemistry, ed. by A. Kasha & B. Pullman, p. 103. New
York: Academic Press.
-
Sonnebom, T. M. (1965). In
Evolving Genes and Proteins, ed. by V. Bryson & H. J. Vogel,
p. 377. New York: Academic Press.
-
Welton, M. G. E. & Pele, S. R.
(1966). Nature, 209, 870.
-
Woese, C. (1965). Proc. Nat.
Acad. Sci., Wash. 54, 1546.
-
Woese, C. R. (1967). The Genetic
Code. New York: Harper & Row.
|

