|
from QuantaMagazine Website
At the dawn of life, simple networks of molecules somehow started to evolve, diversify and become more complex. Researchers have now found clues to how this might have happened by watching RNA molecules
evolve in a test tube. Quanta Magazine
When researchers gave a genetic molecule the ability to replicate, it evolved over time into a complex network of "hosts" and "parasites" that both competed and cooperated to survive...
Over hundreds of hours of replication, a single type of RNA evolved into five different molecular "species" or lineages of hosts and parasites that coexisted in harmony and cooperated to survive, like the beginning of a,
Their experiment, which confirmed previous theoretical findings, showed that molecules with the means to replicate could spontaneously develop complexity through Darwinian evolution,
This was the first and probably most important step toward evolving a complex network of replicators in the lab, said Sijbren Otto, a professor of systems chemistry at the University of Groningen in the Netherlands who was not involved in the study.
Joana Xavier, a computational biologist at University College London, hailed the work by Mizuuchi and his colleagues as a "great proof of concept" for how a minimal system can complexity.
It's "a very significant advance," she said.
Despite the overtones of Frankenstein in that label, his little monster was not green, square-browed, growling or even alive.
It was a synthetic molecule
that filled test tubes with copies of itself.
performed the first demonstration of Darwinian evolution at the molecular level, using an evolving strand
of
viral RNA that he called "the little monster."
The biologist had discovered he could indefinitely replicate it simply by heating and mixing it in the presence of nucleotide building blocks and a polymerizing enzyme called a replicase.
He soon realized, however, that his molecules were getting smaller over time:
Just like living species,
his molecules had started mutating and evolving under the pressure
of natural selection to better survive inside their glass world.
Spiegelman's work inspired decades of further study, much of which was foundational to research on life's origins and provided fuel for the RNA world hypothesis that life sprang from self-replicating RNA molecules.
But those studies left unanswered a crucial question: Could a single molecular replicator evolve into a complex network of multiple replicators?
About a decade ago, when Norikazu Ichihashi was an associate professor of bioinformatic engineering at Osaka University in Japan, he set out to learn the answer by tweaking Spiegelman's test tube world.
Ichihashi and his team developed an RNA molecule that encoded a replicase, which can make copies of RNA.
But for the molecule to translate its own code, the scientists needed to add something more: ribosomes and other gene translation machinery that they borrowed from the common gut bacteria Escherichia coli.
They embedded the machinery inside droplets and added them to a mixture of RNAs and raw materials.
Then came years of tedious mixing and waiting.
Their long-term experiment involved incubating their replication system at 37 degrees Celsius (the temperature of a human body or a hot summer's day), adding new droplets with fresh translation systems, and stirring the mixture to induce replication.
Evolution in Test Tubes
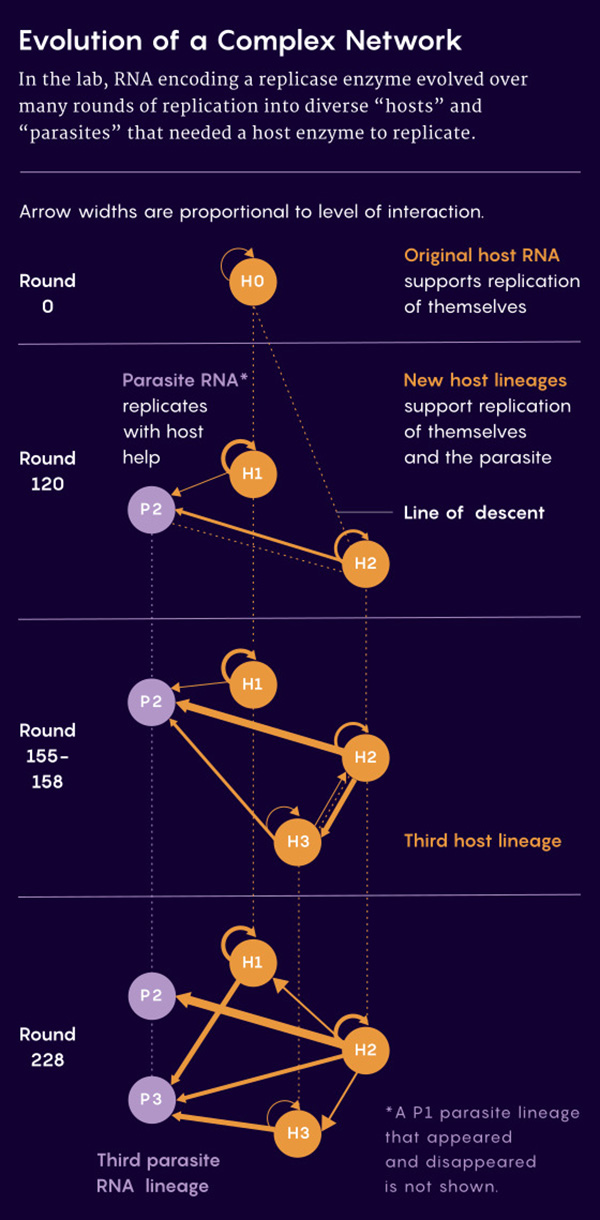
After 215 hours and 43 rounds of replication, the researchers began to see interesting results, which they reported in the Proceedings of the National Academy of Sciences in 2016.
The original RNA had been replaced by lineages of two other RNAs. One, which the researchers described as a "host," could use its own replicase to copy itself, like the original molecules.
The other lineage, a "parasite," needed to borrow the gene expression machinery of the hosts.
When Ichihashi and his colleagues extended the experiment to 120 rounds of replication over 600 hours, they found that the host lineage had split into two separate host lineages, and one of the hosts had evolved two distinct parasites.
But it wasn't just the number of lineages that had increased; so had the complexity of their interactions.
The hosts had acquired mutations that interfered with the ability of the parasites to hijack their replicative resources - but the parasites had also developed mutations that served as a defense against those obstacles.
The hosts and parasites seemed to be coevolving.
The populations of parasites and hosts greatly fluctuated as they competed for the realm in "evolutionary arms races," the scientists reported in 2020 in eLife.
Each RNA lineage transiently rose to dominance, then lost its place to another one.
and lead author of the new study, the work demonstrates how parasites and hosts can push each other to evolve. "Without parasites, this level of diversification
is
probably not possible," he said.
But the researchers kept the experiments going, and,
This stabilization suggested that the lineages were no longer competing to replicate. Instead, they had started to interact as a network and cooperate in a state of quasi-stable coexistence.
Mizuuchi and Ichihashi, who did the experiments with Taro Furubayashi (who was a doctoral student in Ichihashi's lab at the time and is now a research fellow at the University of Tokyo), were floored by the findings, which they reported in Nature Communications in March.
They're just "mere molecules," Mizuuchi said.
Cooperative Parasites do Their Share
Koonin agrees that their findings are striking.
Their,
They watched a single type of molecule replicate and gather mutations under natural selection - but then went further by letting the divergent molecules evolve into a community under one another's influence, just as communities of living cells, animals or people would.
In the process, the researchers explored some of the rules governing what it takes for such complex communities to become stable and enduring.
Some of these results confirmed the predictions of earlier experimental studies of how complexity can arise in viruses, bacteria and eukaryotes, as well as some theoretical work.
A study from Koonin's lab, for instance, also suggested that parasites were inevitable in the emergence of complexity.
Evolutionary pressures that parasites and their hosts place on each other lead both sides to split into new lineages.
A more surprising fundamental principle that emerged was the critical role of cooperation.
The five lineages belonged to different small networks of cooperation, and some were more cooperative than others.
By round 228, for example, one of the three hosts had evolved into a "super cooperator" that could replicate itself and all the other lineages; the other two hosts could each replicate only themselves and one of the parasites.
Scientists have focused on studies of competition in evolution for so long that the role of cooperation,
The cooperation among RNAs was focused entirely on replication in the system that Ichihashi, Mizuuchi and their colleagues observed.
But the researchers hope that it will be possible to coerce the RNAs to evolve a completely different function too, such as a metabolic one, by adjusting the natural selection criteria inside the test tubes.
A Different Destiny
He considers it a good paper but noted that what happened in the laboratory may not translate to what happened at the dawn of life.
Indeed, the scenario in Ichihashi's lab could not reflect what played out at the start of life since the experiments depended on translation machinery from E. coli.
Merrill Sherman Quanta Magazine
But Koonin thinks that if researchers found a way to evolve complexity using truly self-replicating systems of molecules, they would see something very much resembling the networks depicted in the paper.
To Otto, the study suggests that once you've solved the problem of accurate replication with molecules at this level of complexity, they will complexify further:
Carrying on with their work, Ichihashi and his colleagues wanted to see if they could re-create the same sustainable network in a separate experiment, so they extracted samples of the five lineages.
This time, however, they found that while four of the lineages continued to replicate and survive for at least 22 more rounds, the fifth disappeared.
One possibility is that the system was even more complex than the researchers thought, and when they isolated the five lineages, they missed a sixth one critical for the survival of the lineage that disappeared.
With theoretical models, Ichihashi's group confirmed that the four remaining lineages could sustainably and interdependently replicate, and that knocking out any one of the four would lead to the extinction of at least one of the others.
Their simulation also pointed to the counterintuitive discovery that knocking out one of the parasites would lead to the extinction of its host.
Meanwhile, the researchers have continued their main test tube experiments and are waiting to see whether their network will complexify further.
They have also begun similar experiments that use DNA instead of RNA.
|