|

last updated May 21, 2001
from
GodAndScience Website
The existence of large amounts of
non-coding DNA (up to 97% in humans) in the genomes of eukaryotes
has been used as an argument against intelligent design (and the
role of a Creator) and as an argument for the random process of
evolution (1). Two evolutionary theories attempted to
explain the reason for the existence of non-coding DNA.
One theory stated that non-coding DNA
was "junk" that consisted of randomly-produced sequences that had
lost their coding ability or partially duplicated genes that were
non-functional. The second theory stated that non-coding DNA was
"selfish", in that it consisted of DNA that preferentially
replicated more efficiently that coding DNA, even though it provided
no selective advantage (in fact was somewhat detrimental since it
was parasitic).
There have always been problems with
these arguments, which have been ignored by many of those making
these claims.
The main question presented by proponents of the
"junk" or "selfish" DNA theories is,
"Why would a perfect God create
flawed DNA which is primarily composed of useless, non-coding
regions?"
The definitive answer has finally
arrived, although for many years there have been strong suggestions
of what the non-coding DNA is doing in our genomes.
Crytomonad
flagellated
single-celled photosynthetic organism
A recent study has shown that eukaryotic
non-coding DNA (also called "secondary DNA) is functional as
a structural element in the nucleus.
The new study examined the genomes of
the single-celled photosynthetic organisms know as Crytomonads.
These organisms exist as vastly different cell sizes, with the
nucleus being proportional in size to that of the cell. Researchers
discovered that the amount of non-coding DNA was proportional to the
size of the nucleus, suggesting that more non-coding DNA was
structurally required in larger nuclei.
As an added proof, the nucleomorph,
a small piece of DNA contained within the plastid that codes for
itself and photosynthetic function, was not changed in size, despite
changes in cell size and nuclear content. The new study is a
stunning rebuttal to the evolutionary theories that attempt to
discredit design and promote concepts such as "junk" DNA and
"selfish" DNA.
The conclusion according to the authors
is quoted below:
"Furthermore, the present lack of
significant amounts of nucleomorph secondary DNA confirms
that selection can readily eliminate functionless nuclear DNA,
refuting 'selfish' and 'junk' theories of secondary DNA".
(see Beaton, M.J. and T.
Cavalier-Smith. 1999.
Eukaryotic non-coding DNA is functional:
evidence from the differential scaling of cryptomonal genomes.
Proc. R. Soc. Lond. B. 266:2053-2059.)
The answers to the question of "junk"
DNA have been coming in for years, and we now know that the
"junk" is not really junk.
Since "junk" DNA is not really junk,
from now on I will call it "non-coding" DNA. Let's first look
at some of the early studies which indicated that there was some
design behind the non-coding DNA. Initial and subsequent studies
showed there were long areas of non-coding DNA which contained
palindromes, thus maintaining symmetry between complementary strands
(2).
Other studies, examining large regions
of genomes, using statistical techniques borrowed from linguistics,
have shown patterns in the non-coding DNA similar to that seen in
human languages (3).
For example, when you take human
language texts and create a histogram plotting the log of the
frequency of occurrence of words against the log of the rank, the
resulting plot is always linear with a slope of -1 for every human
language. Likewise, when you perform the same plot for coding and
non-coding DNA, the plot for the non-coding DNA exhibits a nearly
perfect linear relationship (much better than that seen for the
coding regions of DNA).
The purpose or function of this "DNA
language" was not determined. Another study showed that DNA contains
large areas with unexplained patterns (4).
Such patterns could not be the result of
random chance as stated by Dr. H. Eugene Stanley (Boston
University),
"it is almost incredible that the
occupant of one site on a gene would somehow influence which
nucleotide shows up even 100,000 bases away."
Scientists have noticed for some time
that eukaryotic genomes consist of large amounts of transposable
and interspersed repetitive elements (TIREs).
A study of insect TIREs showed that the
Lepidopteran Bombyx mori (the silkmoth) exhibits the short
interspersion pattern in which Alu-like TIREs predominate while
Drosophila possesses the long interspersion pattern in which
retroviral-like TIREs are prevalent.
An analysis of these sequences revealed
highly non-random patterns of TIREs in both Bombyx and Drosophila.
These patterns suggested that these sequences were under cellular
regulation rather than useless or selfish junk DNA (5).
Later studies showed that simple, repetitive (gt)n(ga)m DNA
sequences are present in major histocompatibility complex MHC-DRB
genes for many distantly related animals, such as artiodactyla
(large hoofed mammals) and man. Obviously, if these sequences were
truly junk, they would not be expected to have been preserved
through millions of years of evolution.
Gel retardation experiments revealed
that these simple repetitive (gt)n(ga)m sequences bind nuclear
proteins and show characteristics of a specific DNA-protein
interaction (6). What functions these DNA-protein
interactions exhibit has not been determined. However, there are
many other examples of DNA-protein interaction which exhibit
regulatory control of DNA transcription.
Other studies have demonstrated the remarkable similarity of
sequence homology in the T-cell receptor genes of mice and men.
Scientists compared the DNA sequence of nearly 100 kilobases of
contiguous DNA in the C delta to C alpha region of the alpha/delta
T-cell receptor loci (TCRAC/TCRDC) of mouse and man. This analysis,
the largest genomic sequence comparison so far, identified a very
high level of organizational and noncoding sequence
similarity (approximately 71%).
The authors conclude,
"This observation begins to question
the notion that much of the chromosomal non-coding sequence is
junk (7)."
More definitive studies have shown that
non-coding DNA provides structure to DNA so that it can perform many
functions which would be impossible without some form of structure.
One of the readily apparent differences
between prokaryotic and eukaryotic DNA is that eukaryotic DNA is
organized into chromosomes, which is further organized into
chromatin code. This kind of structure does not "just happen" for
DNA - it requires specific design. The coding regions of DNA are
concentrated in the chromosomal regions which are the richest in G
(guanine) and C (cytosine) and seem to correspond to the telomeric
regions of certain chromosome arms (T-bands) (8).
Scientists have genetically modified and
therefore removed a single telomere of one chromosome in yeast cells
(9).
The elimination of the telomere caused
cell cycle arrest (stopping of cell division), indicating that
telomeres help cells to distinguish intact chromosomes from damaged
DNA. In the cells that recovered from the arrest the chromosome was
eventually lost, demonstrating that telomeres are essential for
maintaining chromosome stability.
Therefore, non-coding DNA is
absolutely necessary for chromosomal structure and function.
Studies published in February, 1997,
show that organisms produce special proteins that bind to the
telomeres during DNA replication (10). These proteins are
counted in order to determine how long the telomeric DNA should be,
otherwise the telomere would be shortened with each replication,
eventually resulting in loss of critical genes.
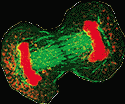 As
you learned in high school, the chromosomes are replicated and
segregated during mitosis (cell division). Complex interactions
occur between the centromeres of chromosomes and the spindles to
which they attach. These centromeres form an integrated protein/DNA
complex, which is required for chromosomal movement during mitosis
(11). As
you learned in high school, the chromosomes are replicated and
segregated during mitosis (cell division). Complex interactions
occur between the centromeres of chromosomes and the spindles to
which they attach. These centromeres form an integrated protein/DNA
complex, which is required for chromosomal movement during mitosis
(11).
What you may not have learned is that
the metaphase chromosomes are dynamically modified in interphase.
In interphase nuclei, orderly transcription and replication
involve highly folded chromosomal domains containing hundreds of
kilobases of DNA.
Specific non-coding DNA sequences within
selected chromosome domains participate in more complex levels of
chromosome folding, and index different genetic compartments for
orderly transcription and replication.
There is also evidence that
three-dimensional chromosome positions within the nucleus contribute
to phenotypic expression. Entire chromosomes are maintained as
discrete, reasonably compact entities in the nucleus, and
heterochromatic coiled domains of several thousand kilobases can
acquire unique three-dimensional positions in differentiated cell
types. This unique structure controls the expression of specific
genes in cells of differentiated cell types (12).
Therefore, non-coding DNA is essential
for differential gene expression seen in the differentiated cell
types seen throughout eukaryotic organisms.
Recent advances have demonstrated that non-protein-coding DNA
provides the structural basis of the metaphase chromosomal banding
pattern. CpG islands, DNA loops, and matrix attachment sites form
the basis of G versus R banding patterns, revealing how non-coding
DNA forms the basis of chromosomal structure (13).
Another study, examining a 2.84 Mb section of the human genome,
showed that microsatellites, tandem repeat sequences abundant in the
genomes of higher eukaryotes, contain reiterating A-rich loci, which
are involved in the higher-order organization of the chromatin
(14).
Other studies have shown satellites
consisting of about 1 million copies of a 221-bp tandem repeat unit
has been localized in the centromeres of 58 of the 64 horse
chromosomes (15). Many hundreds of studies have
implicated mutations in satellites, mini-satellites, and
microsatellites, in diseases which show genetic linkage, including
studies on Crohn's disease, of which I have been part of.
It appears that heterochromatin, composed of what was once thought
to be junk DNA, may have some role in suppression of gene(s) and/or
spreading of inactivation, if genes are embedded within the
heterochromatic region (16).
In a recent study, investigators
examined, through genetic engineering, the relationship between
exon (protein coding DNA) and intron (non-coding DNA)
size in pre-mRNA (messenger RNA, from which protein translation is
accomplished) processing. Exons were placed in vertebrate genes
along with small and large introns.
Both exon and intron size influenced
splicing phenotype, such that when introns were large, large exons
were skipped; when introns were small, the same large exons were
included. These results indicated that non-coding introns can
control the recognition and transcription of exons (protein-coding
DNA) (17). In addition, introns encoded within transfer RNA (tRNA)
genes, which recognize the genetic code on mRNA, code for their
splicing, which allow them to recognize amino acids during the
protein translation process (18).
There is growing evidence that noncoding DNA plays a vital
role in the regulation of gene expression during development
(19). These studies demonstrate that non-coding DNA
regulates development of photoreceptor cells (20), the
reproductive tract (21), and the central nervous system
(22).
Therefore, non-coding DNA regulates
the vital roles of development and embriogenesis.
Some of the non-coding DNA provides proper framing for translation
of proteins. The DNA is, of course, a triplet code, with each
triplet coding for the placement of one amino acid. In order to be
read properly, the reading frame must be properly established. If
the reading frame were shifted by one or two positions in either
direction, the resulting protein would be completely different and
would be "junk" protein. Therefore, the translation framing code is
responsible for correct triplet counting by the ribosome during
protein synthesis (23).
A recent study has shown that genes (as many as five at a time) are
found within the introns of other genes (24).
This kind of arrangement results in the
simultaneous expression of all of these genes during transcription
of the gene in question. Such regulatory control is rather
remarkable, suggesting intelligent designed as opposed to random
chance. Some of the non-coding DNA is loop code for single-stranded
RNA-protein interactions.
The codes are degenerate and
corresponding messages are not only interspersed but actually
overlap, so that some nucleotides belong to several messages
simultaneously. Tandemly repeated sequences frequently considered as
functionless "junk" are found to be grouped into certain classes of
repeat unit lengths, indicating functional involvement of these
sequences.
It is likely these tandem repeats play
the role of weak enhancer-silencers that modulate, in a copy
number-dependent way, the expression of proximal genes.
Well over 700 studies (over 100 in the last year) have demonstrated
the role of non-coding DNA as enhancers for transcription of
proximal genes. These intronic enhancers have been described for:
-
eosinophil-derived neurotoxin (EDN)
and eosinophil cationic protein (ECP) (25)
-
the variable region of the
rearranged immunoglobulin mu (IgM) gene (26)
-
the alpha-globin gene (27)
-
the activin beta A subunit gene
(28)
-
lambda 2 light chain transgenes
(29)
-
Human CYP1B1, a member of the
cytochrome P450 superfamily (30)
-
immunoglobulin heavy chain (IgH)
(31)
-
alcohol dehydrogenase (32)
-
3 alpha-hydroxysteroid
dehydrogenases (33)
-
apolipoprotein A-II (34)
-
beta1,4-N-acetylgalactosaminyltransferase (35)
-
kappa light chain gene (36)
-
Alpha-1 acid glycoprotein
(37)
-
the T-cell receptor beta-chain
(38)
-
2-crystallin (39)
-
1 tubulin gene (40)
-
aldolase B gene (41)
-
and many others...
Another 60+ studies have demonstrated
the role of non-coding DNA as silencers for suppression of
transcription of proximal genes.
The presence of silencer genes has been
shown to down-regulate:
-
the apolipoprotein A-II gene
(42)
-
the osteocalcin gene (43)
-
the 2-crystallin gene (44)
-
the CD4 gene (45)
-
the beta globin gene (46)
-
the gene for the neuron-glia
cell adhesion molecule, Ng-CAM (47)
-
the renin gene (48)
-
the keratin 18 gene (49)
-
the platelet-derived growth
factor A-chain gene (50)
-
and dozens of other genes...
In addition, there are 3' and 5'
untranslated regions (UTR) which regulate translation of proteins.
Certain trans-acting binding proteins bind to the 3' and 5' UTRs of
proximal and distal genes to regulate their translation.
This non-coding DNA has been shown to
regulate:
-
the Lipoprotein Lipase gene
(51)
-
the glucose transporter gene
(52)
-
coxsackie B3 virus (53)
-
the bax-alpha gene (54)
-
glutathione peroxidase and
phospholipid-hydroperoxide glutathione peroxidase genes
(55)
-
the FMR1 gene (56)
-
the c-mos gene (57)
-
the luteinizing hormone/human
chorionic gonadotropin receptor gene (58)
-
the thyrotropin receptor gene
(59)
-
the beta-globin gene (60)
-
the interleukin 1 type I
receptor gene (61)
-
the translation initiation
factor eIF-2 alpha gene (62)
-
the N-methyl-D-aspartate
receptor NR2A subunit gene (63)
-
the catalase gene (64).
In addition, 3'UTRs have been shown to
down-regulate translation of maternal mRNA in oocytes
(65), therefore playing a role in embriogenesis and
development.
Another role for the 3' and 5' UTR is to
regulate the rate of mRNA decay, which has now been shown to be a
precise process dependent on a variety of specific cis-acting
sequences and trans-acting factors (66).
The roles of non-coding DNA are so numerous and pervasive that
evolutionary studies are now looking at these sequences for patterns
of "concerted evolution (67)." In summary, the non-coding
DNA, contrary to statements by evolutionists, is not useless, but
is, in fact, required for genomic functionality, therefore providing
evidence of intelligent design.
The "junk" DNA is really some
rather amazing "junk."
References
1. Moore MJ. 1996. When the junk
isn't junk. Nature 379: 402-403.
2. Ohno S, Yomo T. 1991. The grammatical rule for all DNA: junk
and coding sequences. Electrophoresis 12: 103-108.
Wohr G, Fink T, Assum G. 1996. A palindromic structure in the
pericentromeric region of various human chromosomes. Genome Res
6: 267-279.
3. Flam F. 1994. Hints of a language in junk DNA. Science 266:
1320.
Mantegna RN. et al. 1994. Linguistic features of noncoding DNA
sequences. Physical Reviews Letters 73: 3169-3172.
4. Amato, I. 1992. DNA shows unexplained patterns writ large.
Science 257: 747.
5. von Sternberg RM, Novick GE, Gao GP, Herrera RJ. 1992. Genome
canalization: the coevolution of transposable and interspersed
repetitive elements with single copy DNA. Genetica 86: 215-246.
6. Maueler W, Muller M, Kohne AC, Epplen JT. 1992. A gel
retardation assay system for studying protein binding to simple
repetitive DNA sequences. Electrophoresis 13: 7-10.
7. Koop BF, Hood L. 1994. Striking sequence similarity over
almost 100 kilobases of human and mouse T-cell receptor DNA. Nat
Genet 7: 48-53.
8. Bernardi G. 1991. The human genome: a view from above. Boll
Soc. Ital. Biol. Sper. 67: 459-474.
9. Sandell LL, Zakian VA. 1994. Loss of a yeast telomere:
arrest, recovery, and chromosome loss. Cell 75: 729-739.
10. Barinaga, M. 1997. Cells count proteins to keep their
telomeres in line. Science 275: 928.
Harrington L, McPhail T, Mar V, Zhou W, Oulton R, Bass MB,
Arruda I, Robinson MO. 1997. A mammalian telomerase-associated
protein. Science 275: 973-977.
Marcand S, Gilson E, Shore D. 1997. A protein-counting mechanism
for telomere length regulation in yeast. Science 275: 986-990.
11. Schulman I, Bloom KS. Centromeres: an integrated protein/DNA
complex required for chromosome movement. Annu Rev Cell Biol 7:
311-336.
12. Manuelidis L. A view of interphase chromosomes. Science 250:
1533-1540.
13. Gardiner K. 1995. Human genome organization. Curr. Opin.
Genet Dev. 5: 315-322.
14. Nadir E, Margalit H, Gallily T, Ben-Sasson SA. 1996.
Microsatellite spreading in the human genome: evolutionary
mechanisms and structural implications. Proc. Natl. Acad. Sci. U
S A 93: 6470-6475.
15. Wijers ER, Zijlstra C, Lenstra JA. 1993. Rapid evolution of
horse satellite DNA. Genomics 18: 113-117.
16. Macera MJ, Verma RS, Conte RA, Bialer MG, Klein VR. 1996.
Mechanisms of the origin of a G-positive band within the
secondary constriction region of human chromosome 9. Cytogenet.
Cell Genet 69: 235-239.
17. Sterner DA, Carlo T, Berget SM. 1996. Architectural limits
on split genes. Proc. Natl. Acad. Sci. U S A 93: 15081-15085.
18. Abelson, J. 1992. Recognition of tRNA precursors: a role for
the intron. Science 255: 1390.
19. Ting SJ. 1995. A binary model of repetitive DNA sequence in
Caenorhabditis elegans. DNA Cell Biol. 14: 83-85.
20. Vandendries ER, Johnson D, Reinke R. 1996. Orthodenticle is
required for photoreceptor cell development in the Drosophila
eye. Dev Biol 173: 243-255.
21. Keplinger BL, Rabetoy AL, Cavener DR. 1996. A somatic
reproductive organ enhancer complex activates expression in both
the developing and the mature Drosophila reproductive tract. Dev
Biol 180: 311-323.
22. Kohler J, Schafer-Preuss S, Buttgereit D. 1996. Related
enhancers in the intron of the beta1 tubulin gene of Drosophila
melanogaster are essential for maternal and CNS-specific
expression during embryogenesis. Nucleic Acids Res 24:
2543-2550.
Kallunki P, Jenkinson S, Edelman GM, Jones FS. 1995. Silencer
elements modulate the expression of the gene for the neuron-glia
cell adhesion molecule, Ng-CAM. J Biol Chem 270: 21291-21298.
23. Trifonov, E.N. 1989. The multiple codes of nucleotide
sequences. Bull. Math Biol. 51: 417-432.
24. Waterston, R. and Sulston, J. 1995. The genome of
Caenorhabditis elegans. Proc. Nat. Acad. Sci. USA 92
10836-10840.
25. Handen JS, Rosenberg HF. 1997. Intronic Enhancer Activity of
the Eosinophil-derived Neurotoxin (RNS2) and Eosinophil Cationic
Protein (RNS3) Genes Is Mediated by an NFAT-1 Consensus Binding
Sequence. J Biol. Chem. 272: 1665-1669.
Tiffany HL, Handen JS, Rosenberg HF. 1996. Enhanced expression
of the eosinophil-derived neurotoxin ribonuclease (RNS2) gene
requires interaction between the promoter and intron. J Biol
Chem 271: 12387-12393.
26. Jenuwein T, Forrester WC, Fernandez-Herrero LA, Laible G,
Dull M, Grosschedl R. 1997. Extension of chromatin accessibility
by nuclear matrix attachment regions. Nature 385: 269-272.
Nikolajczyk BS, Nelsen B, Sen R. 1996. Precise alignment of
sites required for mu enhancer activation in B cells. Mol Cell
Biol 16: 4544-4554.
27. Bouhassira EE, Kielman MF, Gilman J, Fabry MF, Suzuka S,
Leone O, Gikas E, Bernini LF, Nagel RL. 1997. Properties of the
mouse alpha-globin HS-26: relationship to HS-40, the major
enhancer of human alpha-globin gene expression. Am J Hematol 54:
30-39.
28. Tanimoto K, Yoshida E, Mita S, Nibu Y, Murakami K, Fukamizu
A. 1996. Human activin betaA gene. Identification of novel 5'
exon, functional promoter, and enhancers. J Biol Chem 271:
32760-32769.
29. Klotz EL, Storb U. 1996. Somatic hypermutation of a lambda 2
transgene under the control of the lambda enhancer or the heavy
chain intron enhancer. J Immunol 157: 4458-4463.
30. Tang YM, Wo YYP, Stewart J, Hawkins AL, Griffin CA, Sutter
TR, Greenlee WF. 1996. Isolation and characterization of the
human cytochrome P450 CYP1B1 gene. J Biol Chem 271: 28324-28330.
31. Gstaiger M, Hovens C, Georgiev O, Knoepfel L, Schaffner W.
1996. BZLF1 (ZEBRA, Zta) protein of Epstein-Barr virus selected
in a yeast one-hybrid system by binding to a consensus site in
the IgH intronic enhancer: a role in immunoglobulin expression?
Biol Chem Hoppe Seyler 377: 669-673.
32. McKenzie RW, Brennan MD. 1996. The two small introns of the
Drosophila affinidisjuncta Adh gene are required for normal
transcription. Nucleic Acids Res 24: 3635-3642.
33. Penning TM. 1996. 3 alpha-hydroxysteroid dehydrogenase:
three dimensional structure and gene regulation. J Endocrinol
150: S175-S187.
34. Bossu JP, Chartier FL, Fruchart JC, Auwerx J, Staels B,
Laine B. 1996. Two regulatory elements of similar structure and
placed in tandem account for the repressive activity of the
first intron of the human apolipoprotein A-II gene. Biochem J
318: 547-553.
35. Furukawa K, Soejima H, Niikawa N, Shiku H. 1996. Genomic
organization and chromosomal assignment of the human beta1,
4-N-acetylgalactosaminyltransferase gene. Identification of
multiple transcription units. J Biol Chem 271: 20836-20844.
36. LaVallee TM, Morrison SL. 1996. Identification and
functional characterization of a highly conserved sequence in
the intron of the kappa light chain gene. Mol Immunol 33:
973-988.
Roque MC, Smith PA, Blasquez VC. 1996. A developmentally
modulated chromatin structure at the mouse immunoglobulin kappa
3' enhancer. Mol Cell Biol 16: 3138-3155.
37. Lee YM, Miau LH, Chang CJ, Lee SC. 1996. Transcriptional
induction of the alpha-1 acid glycoprotein (AGP) gene by
synergistic interaction of two alternative activator forms of
AGP/enhancer-binding protein (C/EBP beta) and NF-kappa B or
Nopp140. Mol Cell Biol 16: 4257-4263.
38. Bories JC, Demengeot J, Davidson L, Alt FW. 1996.
Gene-targeted deletion and replacement mutations of the T-cell
receptor beta-chain enhancer: the role of enhancer elements in
controlling V(D)J recombination accessibility. Proc Natl Acad
Sci U S A 93: 7871-7876.
39. Dirks RP, Kraft HJ, Van Genesen ST, Klok EJ, Pfundt R,
Schoenmakers JG, Lubsen NH. 1996. The cooperation between two
silencers creates an enhancer element that controls both the
lens-preferred and the differentiation stage-specific expression
of the rat beta B2-crystallin gene. Eur J Biochem 239: 23-32.
40. Kohler J, Schafer-Preuss S, Buttgereit D. 1996. Related
enhancers in the intron of the beta1 tubulin gene of Drosophila
melanogaster are essential for maternal and CNS-specific
expression during embryogenesis. Nucleic Acids Res 24:
2543-2550.
41. Sabourin JC, Kern AS, Gregori C, Porteu A, Cywiner C,
Chatelet FP, Kahn A, Pichard AL. 1996. An intronic enhancer
essential for tissue-specific expression of the aldolase B
transgenes. J Biol Chem 271: 3469-3473.
42. Bossu JP, Chartier FL, Fruchart JC, Auwerx J, Staels B,
Laine B. 1996. Two regulatory elements of similar structure and
placed in tandem account for the repressive activity of the
first intron of the human apolipoprotein A-II gene. Biochem J
318: 547-553.
43. Goto K, Heymont JL, Klein-Nulend J, Kronenberg HM, Demay MB.
1996. Identification of an osteoblastic silencer element in the
first intron of the rat osteocalcin gene. Biochemistry 35:
11005-11011.
44. Dirks RP, Kraft HJ, Van Genesen ST, Klok EJ, Pfundt R,
Schoenmakers JG, Lubsen NH. 1996. The cooperation between two
silencers creates an enhancer element that controls both the
lens-preferred and the differentiation stage-specific expression
of the rat beta B2-crystallin gene. Eur J Biochem 239: 23-32.
45. Donda A, Schulz M, Burki K, De Libero G, Uematsu Y. 1996.
Identification and characterization of a human CD4 silencer. Eur
J Immunol 26: 493-500.
Siu G, Wurster AL, Duncan DD, Soliman TM, Hedrick SM. 1994. A
transcriptional silencer controls the developmental expression
of the CD4 gene. EMBO J 13: 3570-3579.
46. Wandersee NJ, Ferris RC, Ginder GD. 1996. Intronic and
flanking sequences are required to silence enhancement of an
embryonic beta-type globin gene. Mol Cell Biol 16: 236-246.
47. Kallunki P, Jenkinson S, Edelman GM, Jones FS. 1995.
Silencer elements modulate the expression of the gene for the
neuron-glia cell adhesion molecule, Ng-CAM. J Biol Chem 270:
21291-21298.
48. Voigtlander T, Ripperger A, Ganten D, Bader M. 1995.
Transcriptional silencer in intron I of the rat renin gene. Adv
Exp Med Biol 377: 285-292.
49. Pankov R, Neznanov N, Umezawa A, Oshima RG. 1994. AP-1, ETS,
and transcriptional silencers regulate retinoic acid-dependent
induction of keratin 18 in embryonic cells. Mol Cell Biol 14:
7744-7757.
50. Wang ZY, Masaharu N, Qiu QQ, Takimoto Y, Deuel TF. 1994. An
S1 nuclease-sensitive region in the first intron of human
platelet-derived growth factor A-chain gene contains a
negatively acting cell type-specific regulatory element. Nucleic
Acids Res 22: 457-464.
51. Ranganathan G, Vu D, Kern PA. 1997. Translational Regulation
of Lipoprotein Lipase by Epinephrine Involves a Trans-acting
Binding Protein Interacting with the 3' Untranslated Region. J
Biol Chem 272: 2515-2519.
52. McGowan KM, Police S, Winslow JB, Pekala PH. 1997. Tumor
necrosis factor-alpha regulation of glucose transporter (GLUT1)
mRNA turnover. Contribution of the 3'-untranslated region of the
GLUT1 message. J Biol Chem 272: 1331-1337.
Boado RJ, Tsukamoto H, Pardridge WM. 1996. Evidence for
translational control elements within the 5'-untranslated region
of GLUT1 glucose transporter mRNA. J Neurochem 67: 1335-1343.
Long SD, Pekala PH. 1996. Regulation of GLUT4 mRNA stability by
tumor necrosis factor-alpha: alterations in both protein binding
to the 3' untranslated region and initiation of translation.
Biochem Biophys Res Commun 220: 949-953.
53. Melchers WJ, Hoenderop JG, Bruins Slot HJ, Pleij CW,
Pilipenko EV, Agol VI, Galama JM. 1997. Kissing of the two
predominant hairpin loops in the coxsackie B virus 3'
untranslated region is the essential structural feature of the
origin of replication required for negative-strand RNA
synthesis. J Virol 71: 686-696.
54. Madison DL, Pfeiffer SE. 1996. Cloning of the 3' end of rat
bax-alpha and corresponding developmental down-regulation in
differentiating primary, cultured oligodendrocytes. Neurosci
Lett 220: 183-186.
55. Bermano G, Arthur JR, Hesketh JE. 1996. Role of the 3'
untranslated region in the regulation of cytosolic glutathione
peroxidase and phospholipid-hydroperoxide glutathione peroxidase
gene expression by selenium supply. Biochem J 320: 891-895.
56. Iber H. 1996. Sequence specific binding of cytosolic
proteins to a 12 nucleotide sequence in the 5' untranslated
region of FMR1 mRNA. Biochim Biophys Acta 1309: 167-173.
57. Steel LF, Telly DL, Leonard J, Rice BA, Monks B, Sawicki JA.
1996. Elements in the murine c-mos messenger RNA 5'-untranslated
region repress translation of downstream coding sequences. Cell
Growth Differ 7: 1415-1424.
58. Lu DL, Menon KM. 1996. 3' untranslated region-mediated
regulation of luteinizing hormone/human chorionic gonadotropin
receptor expression. Biochemistry 35: 12347-12353.
59. Kakinuma A, Chazenbalk G, Filetti S, McLachlan SM, Rapoport
B. 1996. BOTH the 5' and 3' noncoding regions of the thyrotropin
receptor messenger ribonucleic acid influence the level of
receptor protein expression in transfected mammalian cells.
Endocrinology 137: 2664-2669.
60. Russell JE, Liebhaber SA. 1996. The stability of human beta-globin
mRNA is dependent on structural determinants positioned within
its 3' untranslated region. Blood 87: 5314-5323.
61. Ye K, Vannier E, Clark BD, Sims JE, Dinarello CA. 1996.
Three distinct promoters direct transcription of different 5'
untranslated regions of the human interleukin 1 type I receptor:
a possible mechanism for control of translation. Cytokine 8:
421-429.
62. Miyamoto S, Chiorini JA, Urcelay E, Safer B. 1996.
Regulation of gene expression for translation initiation factor
eIF-2 alpha: importance of the 3' untranslated region. Biochem J
315: 791-798.
63. Wood MW, VanDongen HM, VanDongen AM. 1996. The
5'-untranslated region of the N-methyl-D-aspartate receptor NR2A
subunit controls efficiency of translation. J Biol Chem 271:
8115-8120.
64. Reimer DL, Singh SM. 1996. Distinct mRNA-binding proteins
interacting with short repeat sequences of the 3' UTR may be
involved in the post-transcriptional regulation of the mouse
catalase gene, Cas-1. DNA Cell Biol 15: 317-328.
65. Seydoux G. 1996. Mechanisms of translational control in
early development. Curr Opin Genet Dev 6: 555-561.
Walker J, Dale M, Standart N. 1996. Unmasking mRNA in clam
oocytes: role of phosphorylation of a 3' UTR masking
element-binding protein at fertilization. Dev Biol 173: 292-305.
66. Jacobson A, Peltz SW. 1996. Interrelationships of the
pathways of mRNA decay and translation in eukaryotic cells. Annu
Rev Biochem 65: 693-739.
67. Elder JF Jr, Turner BJ. 1995. Concerted evolution of
repetitive DNA sequences in eukaryotes. Q Rev Biol. 70: 297-320.
|


 As
you learned in high school, the chromosomes are replicated and
segregated during mitosis (cell division). Complex interactions
occur between the centromeres of chromosomes and the spindles to
which they attach. These centromeres form an integrated protein/DNA
complex, which is required for chromosomal movement during mitosis
(11).
As
you learned in high school, the chromosomes are replicated and
segregated during mitosis (cell division). Complex interactions
occur between the centromeres of chromosomes and the spindles to
which they attach. These centromeres form an integrated protein/DNA
complex, which is required for chromosomal movement during mitosis
(11).